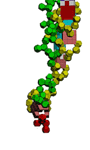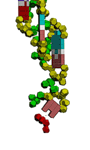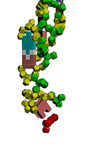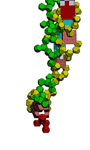Subscribe.Ru : Новости лаборатории Наномир
Выпуск 247
Лаборатория Наномир
Когда реальность открывает тайны,
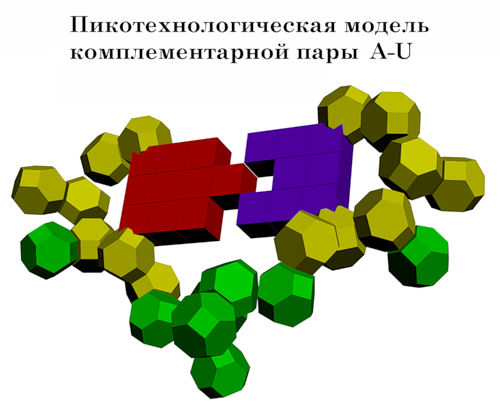
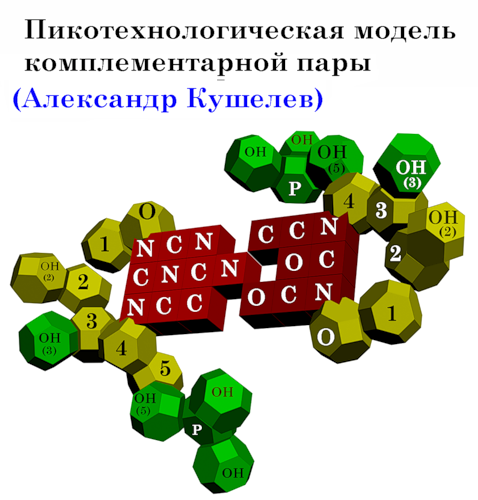
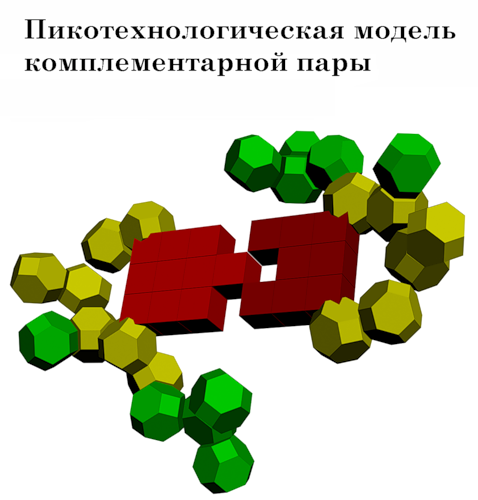
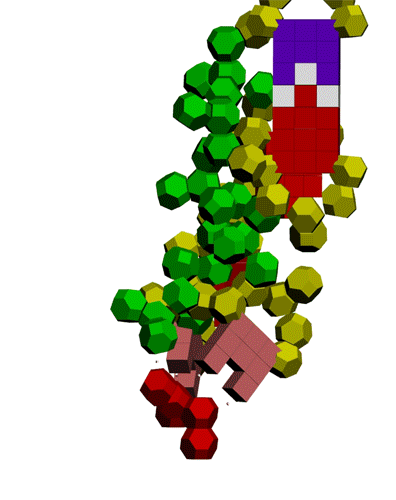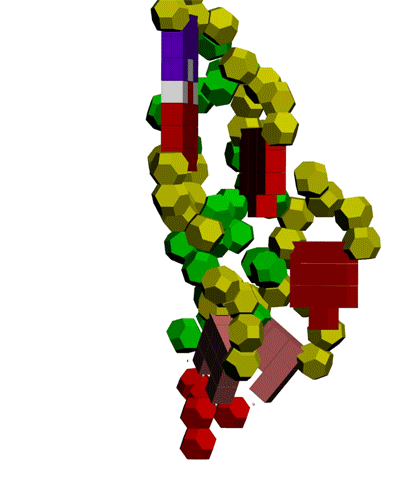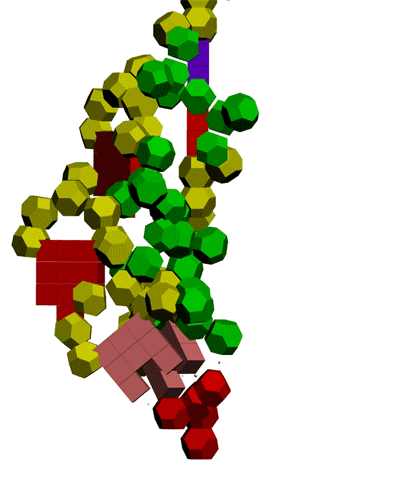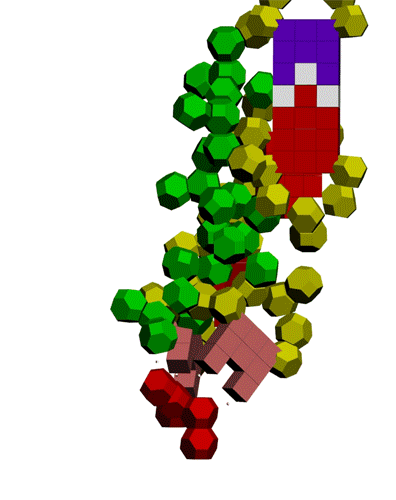
Раскрыта тайна А-минорного взаимодействия!
Кушелев: 2011.06.21
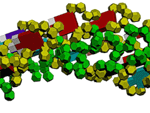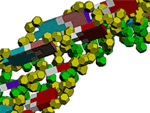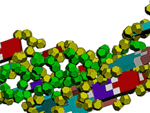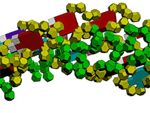
Оказалось,
что параллельно с возникновением водородной связи между азотистым
основанием аденина (А) и третьей CH2-группой рибозы другого нуклеотида
возникает взаимодействие между фосфатными группами этих же нуклеотидов.
Взаимное расположение фосфатных групп напоминает взаимное расположение
радикалов лейцина в лейциновых "молниях-застёжках" белков.
Для
А-минорного взаимодействия характерно, что "А-стопки" в действительности
представляют собой классическую одноцепочечную спираль РНК, ось
симметрии которой совпадает с осью симметрии двухцепочечной спирали, с
которой взаимодействует одноцепочечная. Таким образом, длинные участки с
А-минорным взаимодействием и трёхцепочечная (или трёхнитевая) РНК - это
одна и та же структура
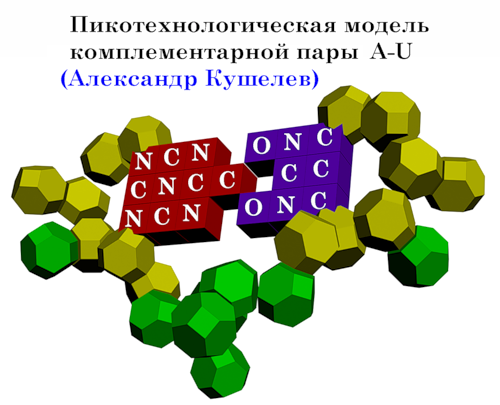
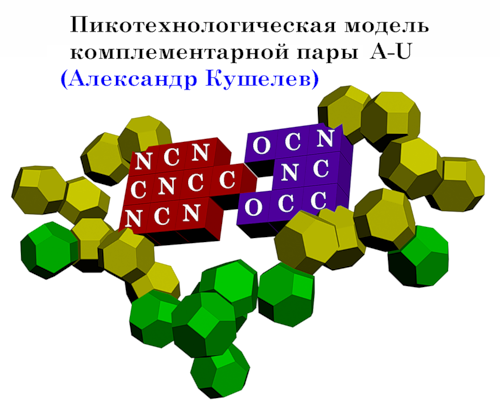
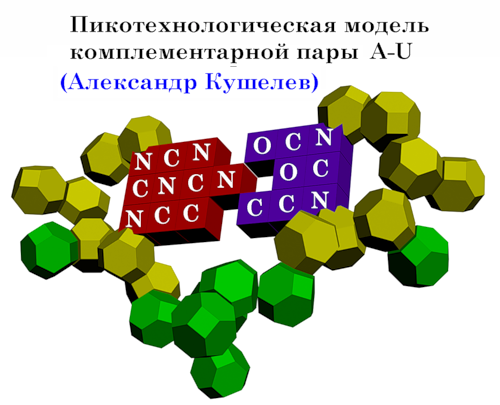
В этом ракурсе хорошо видна диэфирная связь между азотистым основанием
А и третьей СН2-группой рибозы другого нуклеотида.
Кадры крупно можно посмотреть в фотоальбоме "Пикотехнологические
модели"
Маленьким
белым кубиком изображена водородная связь между аденином и второй(?)
группой CH2 рибозы другого нуклеотида. Фосфатные группы аденина показаны
голубым цветом, чтобы легче было отличить от фосфатных групп других
нуклеотидов.
В скрипте добавлена матрица aminor. Для тРНК,
в которой нет А-минорного взаимодействия, эта матрица должна состоять из одних
нулей. В данном скрипте она состоит из единиц, чтобы показать модель
минорного взаимодействия.
Скрипт:
-- Nanoworld Laboratoryhttp://nanoworld.narod.ru /
aa = #(); ax = #(); ay = #(); nuclcol = #(); aminor = #()
newmat = multimaterial name:"MyMultiMat"
numsubs: (14)
Обсудить на форуме.
Научная пикотехнологическая викторина
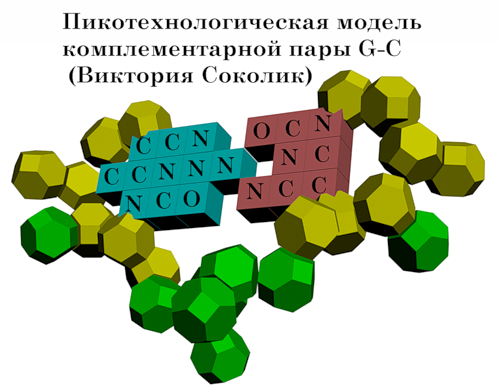
Виктория Соколик обозначила атомы азотистых оснований комплементарной пары G-C
Все
желающие могут иначе обозначить атомы азотистых оснований. Кто знает,
может быть Вам повезёт в это пикотехнологическое "спортлото", и Вы
выиграете Нобелевскую за правильное заполнение "лотерейных билетов"
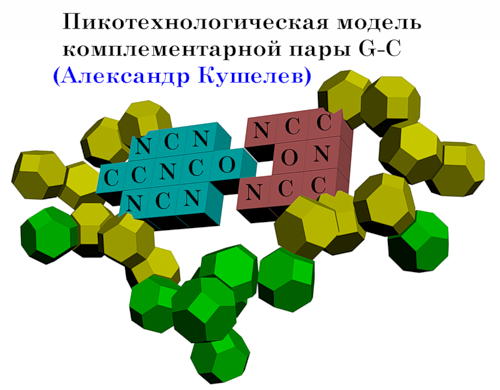
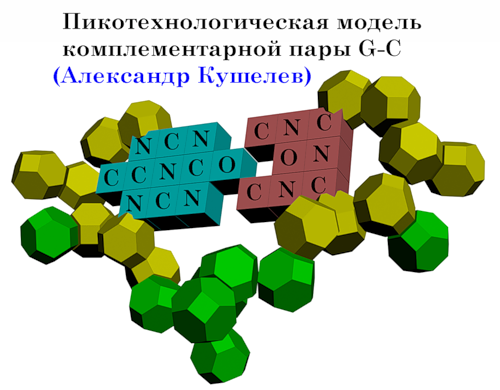
Кушелев: Я тоже решил попытать счастья в научной
пикотехнологической викторине. Предлагаю два варианта заполнения.
Наиболее симметричный вариант может оказаться наиболее устойчивым, т.к.
связь C-N сильнее, чем C-C и N-N.
Карточка пикотехнологической научной викторины N2 A-U
Асимметричная схема расположения атомов наиболее логична при наличии симметричного урацилу псевдоурацила.
Согласно этой классификации, номера атомов рибозы получаются такими:
Желающие могут предложить свой вариант:
Обсудить на форуме
Модель изоакцепторного стебля тРНК
Модель Виктории Соколик
Макромодель тРНК. (Александр Кушелев)
Связь C-N совпадает с осью вращения тРНК
Более
точная пикотехнологическая модель акцепторного стебля с аминокислотой,
удерживаемой четырьмя Ван-дер-Ваальсовыми связями
("макро-взаимодействие").
Кушелев:
Компьютерная модель акцепторного стебля тРНК помогла сделать ещё одно
научное открытие. Оказалось, что азотистые основания, которые держат
аминокислоту четырьмя Ван-дер-Ваальсовыми связями
("макровзаимодействие") кроме этого ещё сдвигают аминокислоту на ось
симметрии тРНК. Таким образом, связь C-N не только параллельна оси
симметрии тРНК, но и вообще совпадает с ней!
Напомню, что общепринятая гипотеза противоречит модельному пикотехнологическому эксперименту по двум параметрам:
1. Угол между направлением C-N и осью симметрии акцепторного стебля ~50 градусов.
В моей модели АСС-конца оба параметра нулевые.
В
этой модели пока не хватает одного нуклеотида (А), но его сравнительно
легко добавить, т.к. он фактически являлся комплементарным к одному из
нуклеотидов (С) до его переворачивания.
Позднее обнаружилось, что аминокислоту
удерживает не 4, а 5 водородных связей. По две на каждое азотистое основание
цитозина и 1 на гидроксил фосфатной группы.
Скрипт:
-- Nanoworld Laboratoryhttp://nanoworld.narod.ru /
aa = #(); ax = #(); ay = #(); aminor = #()
newmat = multimaterial name:"MyMultiMat" numsubs: (14)
"Мозг пикомашины"
Упрощенная (многогранная) пикотехнологическая модель белка инсулина и его зеркальное отражение.
Обсудить
на форуме
Приглашение к сотрудничеству
для людей умеющих самостоятельно мыслить; не просто умных, а мудрых, которые чувствуют, где истина Лаборатория
Наномир готова к любому взаимовыгодному сотрудничеству. У нас
есть сторонники как явные, которые помогают морально и
материально, есть очень много пассивных наблюдателей, есть и ярые
противники, которые используют
любые методы и средства (аморальные и просто преступные), чтобы
уничтожить работу лаборатории и дискредитировать ее.
В одиночку
внедрить технологии, выводящие цивилизацию на новый уровень,
невозможно. Благодаря поддержке множества заинтересованных людей
проделана огромная работа. Ознакомиться с её результатами можно изучив
материал рассылки "Новости лаборатории Наномир". Люди науки могут изучить научные
труды.
Вклад каждого не останется незамеченным в
случае успеха в реализации научных проектов. Результаты совместной
деятельности принадлежат участникам проекта пропорционально
коэффициентам творческого и финансового участия.
В этом году были
куплены рубиновые шарики для эксперимента на сумму ~1000 долл. В
результате было сделано научное открытие, проверена защита
диэлектрических резонаторов от перенапряжения. В этом же году, вероятно,
можно будет создать микроволновую энергетику,
т.к. удалось найти сырьё (рубин #8), из которого сделаны рубиновые
шарики для эксперимента в Дубне.
28 сентября начался эксперимент
по созданию "эликсира вечной молодости". Благодаря первому взносу (в
размере 500 долларов) Золдракса и поддержке других соинвесторов.
Продолжаются переговоры с потенциальными инвесторами по поводу
финансирования этого проекта.
Созданы первые версии пикотехнологии, с помощью которой Александр Кушелев и Виктория Соколик сделали более10 научных открытий.
Сотрудничество
может быть различным:
- участие в научных дискуссиях на форуме (конструктивное)
- совместное создание коммерческого продукта
- поиск инвесторов
- выступить менеджером по продаже готовых коммерческих продуктов
- конструктивные предложения по продвижению идей лаборатории Наномир
- содействие в проведении экспериментов и т.п.
- написание совместных научных статей и т.п.
- материальный вклад (денежный или обеспечение оборудованием и материалами)
Пожалуйста, сообщайте о своем вкладе, чтобы мы зачли Вас как партнера лаборатории Наномир.
+7-926-5101703, +7-903-2003424, +7-916-8265031, Skype: Kushelev2009, mail: kushelev2011@yandex.ru
веб-мани: WM-кошелек R426964799301
![]()