Выпуск 240
Лаборатория Наномир
Когда реальность открывает тайны,
уходят в тень и
меркнут чудеса ...
Причуды ДНК

Экспериментирование с композиционными углами неожиданно привело к "кольцу" из 5 нуклеотидов.
Скрипт:
-- Nanoworld Laboratory
-- Alexander Kushelev
-- Pikotechnological DNA / RNA - model
-- http://nanoworld.narod.ru/
aa = #(); ax = #(); ay = #(); nuclcol = #()
--
adenosine, cytosine, guanine, timidine, uracil, inosine, pseudouracil,
dihydrouracil, methyl inosine, x-circles maximum, w-methyl-2-guanine,
v-methyl-1-guanine
nucl = #("a","c","g","t","u","i","p","d","m","x","w","v")
nuclcolor
=
#([200,0,0],[200,100,100],[0,200,200],[0,0,200],[100,0,200],[200,0,200],[150,0,0],[100,50,0],[0,80,50],[100,100,100])
--*****************************************************************************************************************
seq = #("a","c","c","a","c","c","u","g","c","u","c","a","g","g","c","c","u","u","a","g","c","p","t","g","g","c","c"
,"u","c","d","g","g","a","g","a","g","g","g","p","m","c","g","i","u","u","c","c","c","u","c","w"
,"c","g","c","g","a","d","g","g","c","d","g","a","u","g","c","g","v","u","g","u","g","c","g","g","g")
--*****************************************************************************************************************
--
--*****************************************************************************************************************
--
turn
--
anglecomp1 = #(0,0,0,0,0,0,0,0,0,0, 0,0,0,0,0,0,0,120, 0,
0,-120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0, 0,0,0, 90,0, 90,0,0,0,
0,0,0,0,0,0,0,0,0, 0,120,0,0,0
-- ,0,120, 0, 0,-120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
--
anglecomp2 = #(0,0,0,0,0,0,0,0,0,0,120,0,0,0,0,0,0, 0,-30,-60,
120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0, 0,0,0, -90,0,-90,0,0,0,
0,0,0,0,0,0,0,0,0, 60, 0,0,0,0
-- ,0, 0,-30,-60, 120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
--*****************************************************************************************************************
--
--*****************************************************************************************************************
-- for Victoria Sokolik
turn
anglecomp1
= #(0,0,0,0,0,0,0,0,0,0,
0,0,0,0,0,0,0,20,20,20,20,20,20,20,0,0,0,0,0,0,0,0,0,0,0,0,0, 0,0,0,
90,0, 90,0,0,0, 0,0,0,0,0,0,0,0,0, 0,120,0,0,0
,0,120, 0, 0,-120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
anglecomp2
=
#(0,0,0,0,0,0,0,0,0,0,120,0,0,0,0,0,0,70,70,70,70,70,70,70,0,0,0,0,0,0,0,0,0,0,0,0,0,
0,0,0, -90,0,-90,0,0,0, 0,0,0,0,0,0,0,0,0, 60, 0,0,0,0
,0, 0,-30,-60, 120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
--*****************************************************************************************************************
trnk = torus radius1:0.1 radius2:0.04 segs:3 sides:3 position: [0,0,0] wirecolor:[200,200,200]
Converttomesh trnk
newmat = multimaterial name:"MyMultiMat" numsubs:
(13)
newmat[1].faceted = on
newmat[2].faceted = on
newmat[3].faceted = on
newmat[4].faceted = on
newmat[5].faceted = on
newmat[6].faceted = on
newmat[7].faceted = on
newmat[8].faceted = on
newmat[9].faceted = on
newmat[10].faceted = on
newmat[11].faceted = on
newmat[12].faceted = on
newmat[13].faceted = on
newmat[1].diffuse = (color 200 0 0)
newmat[2].diffuse = (color 200 100 100)
newmat[3].diffuse = (color 0 200 200)
newmat[4].diffuse
= (color 0 0 200)
newmat[5].diffuse = (color 100 0 200)
newmat[6].diffuse = (color 200 0 200)
newmat[7].diffuse = (color 150 0 0)
newmat[8].diffuse = (color 100 50 0)
newmat[9].diffuse = (color 0 80 50)
newmat[10].diffuse = (color 100 100 100)
newmat[11].diffuse = (color 0 200 0)
newmat[12].diffuse = (color 200 200 0)
newmat[13].diffuse = (color 255 255 255)
--
m3 = mesh vertices: #([-2.828,-45.5,-2.828]
,[2.828,-45.5,-2.828],[2.828,-45.5,2.828],[-2.828,-45.5,2.828],[-2.828,-61.5,-2.828],[2.828,-61.5,-2.828]
,[2.828,-61.5,2.828],[-2.828,-61.5,2.828],[-2.828,-53.5,-8.485],[-5.657,-49.5,-5.657],[-8.485,-53.5,-2.828]
,[-5.657,-57.5,-5.657],[8.485,-53.5,2.828],[5.657,-49.5,5.657],[2.828,-53.5,8.485],[5.657,-57.5,5.657]
,[5.657,-49.5,-5.657],[2.828,-53.5,-8.485],[5.657,-57.5,-5.657],[8.485,-53.5,-2.828],[-5.657,-49.5,5.657]
,[-8.485,-53.5,2.828],[-5.657,-57.5,5.657],[-2.828,-53.5,8.485]) \
faces: #([1,3,2],[1,4,3],[5,6,7],[5,7,8],[9,11,10],[9,12,11],[13,14,15],[13,15,16],[17,19,18],[17,20,19],[21,22,23],[21,23,24]
,[1,2,17],[17,18,9],[9,10,1],[1,17,9],[10,22,4],[4,1,10],[10,11,22],[22,21,4],[11,5,23],[23,22,11],[11,12,5],[5,8,23]
,[12,18,6],[6,5,12],[12,9,18],[18,19,6],[8,16,24],[24,23,8],[16,15,24],[8,7,16],[15,3,21],[21,24,15],[15,14,3]
,[3,4,21],[14,20,2],[2,3,14],[14,13,20],[20,17,2],[13,7,19],[19,20,13],[13,16,7],[7,6,19]) \
wirecolor: [255,255,0]
m3.material = newmat[12]
m4 = copy m3 wirecolor: [255,255,0]
m4.material = newmat[12]
m4.pivot = [0,-40,0]
rotate m4 -120 [0,0,1]
move m4 [-7.5,-0.5,0]
m6 = copy m3 wirecolor: [255,255,0]
m6.material = newmat[12]
m13 = copy m3 wirecolor: [255,255,0]
m13.material = newmat[12]
m13.pivot = [0,-66,0]
rotate m13 -72 [1,0,0]
m14 = copy m13 wirecolor: [0,255,0]
m14.material = newmat[11]
g1 = group #(m6, m14)
g1.pivot = [0,-40,0]
rotate g1 -45 [0,0,1]
move g1 [-5.5,2.5,0]
ungroup g1
m5 = copy m6 wirecolor: [255,255,0]
m5.material = newmat[12]
m7 = copy m4 wirecolor:
[255,255,0]
m7.material = newmat[12]
g2 = group #(m5, m7)
g2.pivot = [0,-40,0]
rotate g2 180 [0,1,0]
ungroup g2
m8 = mesh vertices: #([-4,0,-8],[0,-4,-8],[4,0,-8],[0,4,-8],[-4,0,8],[0,-4,8],[4,0,8],[0,4,8]
,[-8,-4,0],[-8,0,-4],[-8,4,0],[-8,0,4],[8,-4,0],[8,0,-4],[8,4,0],[8,0,4]
,[0,-8,-4],[-4,-8,0],[0,-8,4],[4,-8,0],[0,8,-4],[-4,8,0],[0,8,4],[4,8,0]) \
faces: #([1,3,2],[1,4,3],[5,6,7],[5,7,8],[9,11,10],[9,12,11],[13,14,15],[13,15,16],[17,19,18],[17,20,19],[21,22,23],[21,23,24]
,[1,2,17],[17,18,9],[9,10,1],[1,17,9],[10,22,4],[4,1,10],[10,11,22],[22,21,4],[11,5,23],[23,22,11],[11,12,5],[5,8,23]
,[12,18,6],[6,5,12],[12,9,18],[18,19,6],[8,16,24],[24,23,8],[16,15,24],[8,7,16],[15,3,21],[21,24,15],[15,14,3]
,[3,4,21],[14,20,2],[2,3,14],[14,13,20],[20,17,2],[13,7,19],[19,20,13],[13,16,7],[7,6,19]) \
wirecolor: [0,255,0]
m8.material = newmat[11]
rotate m8 45 [1,0,0]
rotate m8 90 [0,0,1]
move m8 [-37.5,0,0]
m9 = copy m8 wirecolor: [0,255,0]
m9.material
= newmat[11]
m9.pivot = [-37.5,-13,-0.5]
rotate m9 72 [1,0,0]
m10 = copy m8 wirecolor: [0,255,0]
m10.material = newmat[11]
m10.pivot = [-37.5,-13,-0.5]
rotate m10 144 [1,0,0]
m11 = copy m8 wirecolor: [0,255,0]
m11.material = newmat[11]
m11.pivot = [-37.5,-13,-0.5]
rotate m11 18 [1,0,0]
rotate m11 90 [0,0,1]
rotate m11 180 [1,0,0]
rotate m11 -18 [1,0,0]
move m11 [0,7,24]
m12 = copy m9 wirecolor: [255,255,0]
m12.material = newmat[12]
m12.pivot = [-37.5,-23.5,15]
rotate m12 -120 [0,0,1]
attach m3 m4
attach m5 m7
attach m6 m13
attach m3 m5
attach m3 m6
attach m8 m9
attach m8 m11
attach m3 m10
attach m12 m14
attach m8 m12
--
for k = 18 to 22 do(
--
adenosine, cytosine, guanine, timidine, uracil, inosine, pseudouracil,
dihydrouracil, methyl inosine, x-circles maximum, w-methyl-2-guanine,
v-methyl-1-guanine
if (seq[k] == nucl[10]) then (ai =
#(1,1,1,1,1,1,1,1,1,1,1,1,1,1,1); axi = #(1,1,1,1,1,1,1,1); ayi =
#(1,1,1,1,1,1,1,1,1,1,1,1,1,1,1); nuclcol = nuclcolor[10]; nc=10)
if
(seq[k] == nucl[1]) then (ai = #(0,0,0,1,1,1,1,1,1,1,1,1,0,1,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol =
nuclcolor[1]; nc=1)
if (seq[k] == nucl[2]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol = nuclcolor[2]; nc=2)
if
(seq[k] == nucl[3]) then (ai = #(0,1,0,1,1,1,1,1,1,1,1,1,0,1,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,0,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol =
nuclcolor[3]; nc=3)
if (seq[k] == nucl[4]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol = nuclcolor[4]; nc=4)
if
(seq[k] == nucl[5]) then (ai = #(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol =
nuclcolor[5]; nc=5)
if (seq[k] == nucl[6]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,1,1,0,1,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol = nuclcolor[6]; nc=6)
if
(seq[k] == nucl[7]) then (ai = #(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol =
nuclcolor[7]; nc=7)
if (seq[k] == nucl[8]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol = nuclcolor[8]; nc=8)
if
(seq[k] == nucl[9]) then (ai = #(0,0,0,1,1,1,1,1,1,1,1,1,0,1,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol =
nuclcolor[9]; nc=9)
--
element1 = box length:1 width:1 height:1 position:[0,0,-0.5] wirecolor: nuclcol
Converttomesh element1
for i = 1 to 3 do for j = 1 to 5 do (n = i + 3*(j-1)
if ai[n] > 0 then (aa[n] = box length:10 width:10 height:10 position:[10*(i-2),10*(j-5),10*0.5-10] wirecolor: nuclcol
select #(aa[n]); macros.run "Modifier stack" "convert_to_Mesh"
attach element1 aa[n]
element1.material = newmat[nc]))
element2 = copy m8
element3 = copy m3
attach element1 element3
-- first vector
gr1 = group #(element1,trnk)
gr1.pivot = [-37.451, -2.887, 14.138]
rotate gr1 anglecomp1[k] [0, -27.384, -9.84]
ungroup gr1
-- second vector
gr2 = #(element1, element2, trnk)
gr2.pivot = [-43.391, -10.855, 7.631]
rotate gr2 anglecomp2[k] [-11.880, -2.245, -8.094]
ungroup gr2
-- translation vector
attach trnk element1
attach trnk element2
move trnk [0,-50, 0]
trnk.pivot
= [-60,0,0]
-- translation angles
rotate trnk 36 [0, 1, 0]
-- rotate trnk 35 [0, 0, 1]
)
--
delete m3
delete m8
trnk.rotation.controller[2].controller.value = 0
-- animate on
-- at time 100 trnk.rotation.controller[2].controller.value = 360
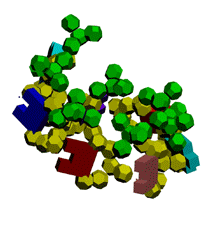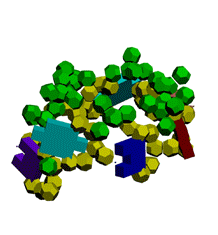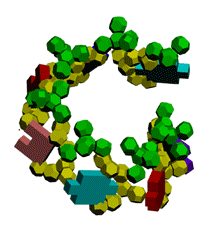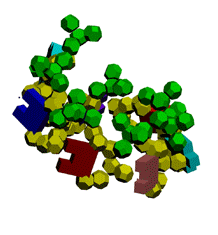
Кушелев: Ну вот Вам композиционные углы, как Вы того и хотели. Получается спиральный виток из 7 нуклеотидов.
Первые композиционные углы по 14 градусов, а вторые - по 50. Но эта модель неустойчивого состояния, из которого петля перейдёт в устойчивое. Одно из устойчивых для каждой (акцепторной, псевдоуридиловой и антикодоновой) я показал на пластмассовых моделях. И пока не дождался от Вас конкретной критики...
Скрипт:
-- Nanoworld Laboratory
-- Alexander Kushelev
-- Pikotechnological DNA / RNA - model
-- http://nanoworld.narod.ru/
aa = #(); ax = #(); ay = #(); nuclcol = #()
--
adenosine, cytosine, guanine, timidine, uracil, inosine, pseudouracil,
dihydrouracil, methyl inosine, x-circles maximum, w-methyl-2-guanine,
v-methyl-1-guanine
nucl = #("a","c","g","t","u","i","p","d","m","x","w","v")
nuclcolor
=
#([200,0,0],[200,100,100],[0,200,200],[0,0,200],[100,0,200],[200,0,200],[150,0,0],[100,50,0],[0,80,50],[100,100,100])
--*****************************************************************************************************************
seq = #("a","c","c","a","c","c","u","g","c","u","c","a","g","g","c","c","u","u","a","g","c","p","t","g","g","c","c"
,"u","c","d","g","g","a","g","a","g","g","g","p","m","c","g","i","u","u","c","c","c","u","c","w"
,"c","g","c","g","a","d","g","g","c","d","g","a","u","g","c","g","v","u","g","u","g","c","g","g","g")
--*****************************************************************************************************************
--
--*****************************************************************************************************************
--
turn
--
anglecomp1 = #(0,0,0,0,0,0,0,0,0,0, 0,0,0,0,0,0,0,120, 0,
0,-120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0, 0,0,0, 90,0, 90,0,0,0,
0,0,0,0,0,0,0,0,0, 0,120,0,0,0
-- ,0,120, 0, 0,-120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
--
anglecomp2 = #(0,0,0,0,0,0,0,0,0,0,120,0,0,0,0,0,0, 0,-30,-60,
120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0, 0,0,0, -90,0,-90,0,0,0,
0,0,0,0,0,0,0,0,0, 60, 0,0,0,0
-- ,0, 0,-30,-60, 120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
--*****************************************************************************************************************
--
--*****************************************************************************************************************
-- for Victoria Sokolik
turn
anglecomp1
= #(0,0,0,0,0,0,0,0,0,0,
0,0,0,0,0,0,0,14,14,14,14,14,14,14,0,0,0,0,0,0,0,0,0,0,0,0,0, 0,0,0,
90,0, 90,0,0,0, 0,0,0,0,0,0,0,0,0, 0,120,0,0,0
,0,120, 0, 0,-120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
anglecomp2
=
#(0,0,0,0,0,0,0,0,0,0,120,0,0,0,0,0,0,50,50,50,50,50,50,50,0,0,0,0,0,0,0,0,0,0,0,0,0,
0,0,0, -90,0,-90,0,0,0, 0,0,0,0,0,0,0,0,0, 60, 0,0,0,0
,0, 0,-30,-60, 120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
--*****************************************************************************************************************
trnk = torus radius1:0.1 radius2:0.04 segs:3 sides:3 position: [0,0,0] wirecolor:[200,200,200]
Converttomesh trnk
newmat = multimaterial name:"MyMultiMat" numsubs:
(13)
newmat[1].faceted = on
newmat[2].faceted = on
newmat[3].faceted = on
newmat[4].faceted = on
newmat[5].faceted = on
newmat[6].faceted = on
newmat[7].faceted = on
newmat[8].faceted = on
newmat[9].faceted = on
newmat[10].faceted = on
newmat[11].faceted = on
newmat[12].faceted = on
newmat[13].faceted = on
newmat[1].diffuse = (color 200 0 0)
newmat[2].diffuse = (color 200 100 100)
newmat[3].diffuse = (color 0 200 200)
newmat[4].diffuse
= (color 0 0 200)
newmat[5].diffuse = (color 100 0 200)
newmat[6].diffuse = (color 200 0 200)
newmat[7].diffuse = (color 150 0 0)
newmat[8].diffuse = (color 100 50 0)
newmat[9].diffuse = (color 0 80 50)
newmat[10].diffuse = (color 100 100 100)
newmat[11].diffuse = (color 0 200 0)
newmat[12].diffuse = (color 200 200 0)
newmat[13].diffuse = (color 255 255 255)
--
m3 = mesh vertices: #([-2.828,-45.5,-2.828]
,[2.828,-45.5,-2.828],[2.828,-45.5,2.828],[-2.828,-45.5,2.828],[-2.828,-61.5,-2.828],[2.828,-61.5,-2.828]
,[2.828,-61.5,2.828],[-2.828,-61.5,2.828],[-2.828,-53.5,-8.485],[-5.657,-49.5,-5.657],[-8.485,-53.5,-2.828]
,[-5.657,-57.5,-5.657],[8.485,-53.5,2.828],[5.657,-49.5,5.657],[2.828,-53.5,8.485],[5.657,-57.5,5.657]
,[5.657,-49.5,-5.657],[2.828,-53.5,-8.485],[5.657,-57.5,-5.657],[8.485,-53.5,-2.828],[-5.657,-49.5,5.657]
,[-8.485,-53.5,2.828],[-5.657,-57.5,5.657],[-2.828,-53.5,8.485]) \
faces: #([1,3,2],[1,4,3],[5,6,7],[5,7,8],[9,11,10],[9,12,11],[13,14,15],[13,15,16],[17,19,18],[17,20,19],[21,22,23],[21,23,24]
,[1,2,17],[17,18,9],[9,10,1],[1,17,9],[10,22,4],[4,1,10],[10,11,22],[22,21,4],[11,5,23],[23,22,11],[11,12,5],[5,8,23]
,[12,18,6],[6,5,12],[12,9,18],[18,19,6],[8,16,24],[24,23,8],[16,15,24],[8,7,16],[15,3,21],[21,24,15],[15,14,3]
,[3,4,21],[14,20,2],[2,3,14],[14,13,20],[20,17,2],[13,7,19],[19,20,13],[13,16,7],[7,6,19]) \
wirecolor: [255,255,0]
m3.material = newmat[12]
m4 = copy m3 wirecolor: [255,255,0]
m4.material = newmat[12]
m4.pivot = [0,-40,0]
rotate m4 -120 [0,0,1]
move m4 [-7.5,-0.5,0]
m6 = copy m3 wirecolor: [255,255,0]
m6.material = newmat[12]
m13 = copy m3 wirecolor: [255,255,0]
m13.material = newmat[12]
m13.pivot = [0,-66,0]
rotate m13 -72 [1,0,0]
m14 = copy m13 wirecolor: [0,255,0]
m14.material = newmat[11]
g1 = group #(m6, m14)
g1.pivot = [0,-40,0]
rotate g1 -45 [0,0,1]
move g1 [-5.5,2.5,0]
ungroup g1
m5 = copy m6 wirecolor: [255,255,0]
m5.material = newmat[12]
m7 = copy m4 wirecolor:
[255,255,0]
m7.material = newmat[12]
g2 = group #(m5, m7)
g2.pivot = [0,-40,0]
rotate g2 180 [0,1,0]
ungroup g2
m8 = mesh vertices: #([-4,0,-8],[0,-4,-8],[4,0,-8],[0,4,-8],[-4,0,8],[0,-4,8],[4,0,8],[0,4,8]
,[-8,-4,0],[-8,0,-4],[-8,4,0],[-8,0,4],[8,-4,0],[8,0,-4],[8,4,0],[8,0,4]
,[0,-8,-4],[-4,-8,0],[0,-8,4],[4,-8,0],[0,8,-4],[-4,8,0],[0,8,4],[4,8,0]) \
faces: #([1,3,2],[1,4,3],[5,6,7],[5,7,8],[9,11,10],[9,12,11],[13,14,15],[13,15,16],[17,19,18],[17,20,19],[21,22,23],[21,23,24]
,[1,2,17],[17,18,9],[9,10,1],[1,17,9],[10,22,4],[4,1,10],[10,11,22],[22,21,4],[11,5,23],[23,22,11],[11,12,5],[5,8,23]
,[12,18,6],[6,5,12],[12,9,18],[18,19,6],[8,16,24],[24,23,8],[16,15,24],[8,7,16],[15,3,21],[21,24,15],[15,14,3]
,[3,4,21],[14,20,2],[2,3,14],[14,13,20],[20,17,2],[13,7,19],[19,20,13],[13,16,7],[7,6,19]) \
wirecolor: [0,255,0]
m8.material = newmat[11]
rotate m8 45 [1,0,0]
rotate m8 90 [0,0,1]
move m8 [-37.5,0,0]
m9 = copy m8 wirecolor: [0,255,0]
m9.material
= newmat[11]
m9.pivot = [-37.5,-13,-0.5]
rotate m9 72 [1,0,0]
m10 = copy m8 wirecolor: [0,255,0]
m10.material = newmat[11]
m10.pivot = [-37.5,-13,-0.5]
rotate m10 144 [1,0,0]
m11 = copy m8 wirecolor: [0,255,0]
m11.material = newmat[11]
m11.pivot = [-37.5,-13,-0.5]
rotate m11 18 [1,0,0]
rotate m11 90 [0,0,1]
rotate m11 180 [1,0,0]
rotate m11 -18 [1,0,0]
move m11 [0,7,24]
m12 = copy m9 wirecolor: [255,255,0]
m12.material = newmat[12]
m12.pivot = [-37.5,-23.5,15]
rotate m12 -120 [0,0,1]
attach m3 m4
attach m5 m7
attach m6 m13
attach m3 m5
attach m3 m6
attach m8 m9
attach m8 m11
attach m3 m10
attach m12 m14
attach m8 m12
--
for k = 18 to 24 do(
--
adenosine, cytosine, guanine, timidine, uracil, inosine, pseudouracil,
dihydrouracil, methyl inosine, x-circles maximum, w-methyl-2-guanine,
v-methyl-1-guanine
if (seq[k] == nucl[10]) then (ai =
#(1,1,1,1,1,1,1,1,1,1,1,1,1,1,1); axi = #(1,1,1,1,1,1,1,1); ayi =
#(1,1,1,1,1,1,1,1,1,1,1,1,1,1,1); nuclcol = nuclcolor[10]; nc=10)
if
(seq[k] == nucl[1]) then (ai = #(0,0,0,1,1,1,1,1,1,1,1,1,0,1,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol =
nuclcolor[1]; nc=1)
if (seq[k] == nucl[2]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol = nuclcolor[2]; nc=2)
if
(seq[k] == nucl[3]) then (ai = #(0,1,0,1,1,1,1,1,1,1,1,1,0,1,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,0,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol =
nuclcolor[3]; nc=3)
if (seq[k] == nucl[4]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol = nuclcolor[4]; nc=4)
if
(seq[k] == nucl[5]) then (ai = #(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol =
nuclcolor[5]; nc=5)
if (seq[k] == nucl[6]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,1,1,0,1,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol = nuclcolor[6]; nc=6)
if
(seq[k] == nucl[7]) then (ai = #(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol =
nuclcolor[7]; nc=7)
if (seq[k] == nucl[8]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol = nuclcolor[8]; nc=8)
if
(seq[k] == nucl[9]) then (ai = #(0,0,0,1,1,1,1,1,1,1,1,1,0,1,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol =
nuclcolor[9]; nc=9)
--
element1 = box length:1 width:1 height:1 position:[0,0,-0.5] wirecolor: nuclcol
Converttomesh element1
for i = 1 to 3 do for j = 1 to 5 do (n = i + 3*(j-1)
if ai[n] > 0 then (aa[n] = box length:10 width:10 height:10 position:[10*(i-2),10*(j-5),10*0.5-10] wirecolor: nuclcol
select #(aa[n]); macros.run "Modifier stack" "convert_to_Mesh"
attach element1 aa[n]
element1.material = newmat[nc]))
element2 = copy m8
element3 = copy m3
attach element1 element3
-- first vector
gr1 = group #(element1,trnk)
gr1.pivot = [-37.451, -2.887, 14.138]
rotate gr1 anglecomp1[k] [0, -27.384, -9.84]
ungroup gr1
-- second vector
gr2 = #(element1, element2, trnk)
gr2.pivot = [-43.391, -10.855, 7.631]
rotate gr2 anglecomp2[k] [-11.880, -2.245, -8.094]
ungroup gr2
-- translation vector
attach trnk element1
attach trnk element2
move trnk [0,-50, 0]
trnk.pivot
= [-60,0,0]
-- translation angles
rotate trnk 36 [0, 1, 0]
-- rotate trnk 35 [0, 0, 1]
)
--
delete m3
delete m8
trnk.rotation.controller[2].controller.value = 0
-- animate on
-- at time 100 trnk.rotation.controller[2].controller.value = 360
Пикотехнология трансляции
Кушелев: Сегодня, 2011-05-29, удалось, наконец, отъюстировать вручную модель антикодоновой петли тРНК.
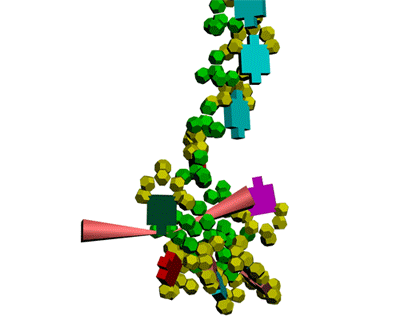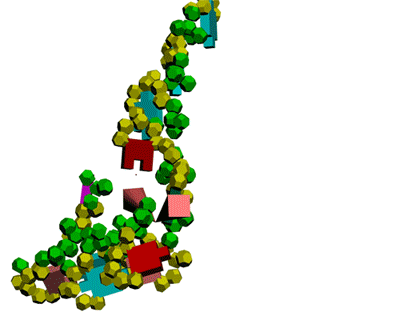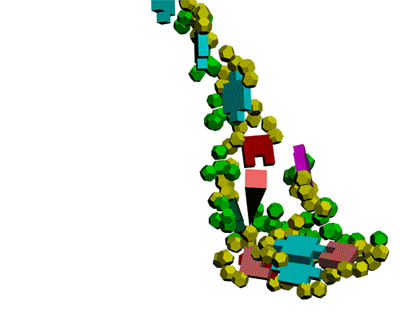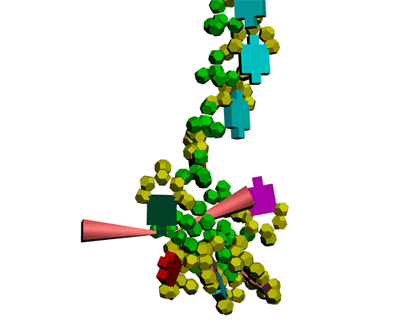
Метилинозин
срезает продольную диэфирную связь, оставляя триплет иРНК на
антикодоновой петле тРНК. тРНК продолжает по инерции вращаться, пока
третий нуклеотид кодона иРНК не встретит комплементарное основание на
сайте рибосомы.
Третий нуклеотид триплета терминирует угол поворота
тРНК в рибосоме. После этого аминокислота устанавливается в растующую
бековую молекулу под нужным углом.
Полный набор виртуальных объектов (фрагмент тРНК, триплет иРНК и две пирамидки-указатели содержатся в файле: http://nanoworld2003.narod.ru/2011052901.max
Скрипт:
-- Nanoworld Laboratory
-- Alexander Kushelev
-- Pikotechnological DNA / RNA - model
-- http://nanoworld.narod.ru/
aa = #(); ax = #(); ay =
#(); nuclcol = #()
--
adenosine, cytosine, guanine, timidine, uracil, inosine, pseudouracil,
dihydrouracil, methyl inosine, x-circles maximum, w-methyl-2-guanine,
v-methyl-1-guanine
nucl = #("a","c","g","t","u","i","p","d","m","x","w","v")
nuclcolor
=
#([200,0,0],[200,100,100],[0,200,200],[0,0,200],[100,0,200],[200,0,200],[150,0,0],[100,50,0],[0,80,50],[100,100,100])
--*****************************************************************************************************************
seq = #("a","c","c","a","c","c","u","g","c","u","c","a","g","g","c","c","u","u","a","g","c","p","t","g","g","c","c"
,"u","c","d","g","g","a","g","a","g","g","g","p","m","c","g","i","u","u","c","c","c","u","c","w"
,"c","g","c","g","a","d","g","g","c","d","g","a","u","g","c","g","v","u","g","u","g","c","g","g","g")
--*****************************************************************************************************************
--
turn
anglecomp1
= #(0,0,0,0,0,0,0,0,0,0, 0,0,0,0,0,0,0,120, 0,
0,-120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,-20,-40, 40,0,-30,0,0,0,0,
0,0,0,0,0,0,0,0,0, 0,120,0,0,0
,0,120, 0, 0,-120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
anglecomp2
= #(0,0,0,0,0,0,0,0,0,0,120,0,0,0,0,0,0, 0,-30,-60,
120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0, 0, 0,-120,0, 120,0,0,0,0,
0,0,0,0,0,0,0,0,0, 60, 0,0,0,0
,0, 0,-30,-60, 120,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0)
--*****************************************************************************************************************
trnk = torus radius1:0.1 radius2:0.04 segs:3 sides:3 position: [0,0,0] wirecolor:[200,200,200]
Converttomesh trnk
newmat = multimaterial name:"MyMultiMat" numsubs: (13)
newmat[1].faceted = on
newmat[2].faceted
= on
newmat[3].faceted = on
newmat[4].faceted = on
newmat[5].faceted = on
newmat[6].faceted = on
newmat[7].faceted = on
newmat[8].faceted = on
newmat[9].faceted = on
newmat[10].faceted = on
newmat[11].faceted = on
newmat[12].faceted = on
newmat[13].faceted = on
newmat[1].diffuse = (color 200 0 0)
newmat[2].diffuse = (color 200 100 100)
newmat[3].diffuse = (color 0 200 200)
newmat[4].diffuse = (color 0 0 200)
newmat[5].diffuse = (color
100 0 200)
newmat[6].diffuse = (color 200 0 200)
newmat[7].diffuse = (color 150 0 0)
newmat[8].diffuse = (color 100 50 0)
newmat[9].diffuse = (color 0 80 50)
newmat[10].diffuse = (color 100 100 100)
newmat[11].diffuse = (color 0 200 0)
newmat[12].diffuse = (color 200 200 0)
newmat[13].diffuse = (color 255 255 255)
--
m3 = mesh vertices: #([-2.828,-45.5,-2.828]
,[2.828,-45.5,-2.828],[2.828,-45.5,2.828],[-2.828,-45.5,2.828],[-2.828,-61.5,-2.828],[2.828,-61.5,-2.828]
,[2.828,-61.5,2.828],[-2.828,-61.5,2.828],[-2.828,-53.5,-8.485],[-5.657,-49.5,-5.657],[-8.485,-53.5,-2.828]
,[-5.657,-57.5,-5.657],[8.485,-53.5,2.828],[5.657,-49.5,5.657],[2.828,-53.5,8.485],[5.657,-57.5,5.657]
,[5.657,-49.5,-5.657],[2.828,-53.5,-8.485],[5.657,-57.5,-5.657],[8.485,-53.5,-2.828],[-5.657,-49.5,5.657]
,[-8.485,-53.5,2.828],[-5.657,-57.5,5.657],[-2.828,-53.5,8.485]) \
faces: #([1,3,2],[1,4,3],[5,6,7],[5,7,8],[9,11,10],[9,12,11],[13,14,15],[13,15,16],[17,19,18],[17,20,19],[21,22,23],[21,23,24]
,[1,2,17],[17,18,9],[9,10,1],[1,17,9],[10,22,4],[4,1,10],[10,11,22],[22,21,4],[11,5,23],[23,22,11],[11,12,5],[5,8,23]
,[12,18,6],[6,5,12],[12,9,18],[18,19,6],[8,16,24],[24,23,8],[16,15,24],[8,7,16],[15,3,21],[21,24,15],[15,14,3]
,[3,4,21],[14,20,2],[2,3,14],[14,13,20],[20,17,2],[13,7,19],[19,20,13],[13,16,7],[7,6,19]) \
wirecolor: [255,255,0]
m3.material = newmat[12]
m4 = copy m3 wirecolor: [255,255,0]
m4.material = newmat[12]
m4.pivot = [0,-40,0]
rotate m4 -120 [0,0,1]
move m4 [-7.5,-0.5,0]
m6 = copy m3 wirecolor: [255,255,0]
m6.material = newmat[12]
m13 = copy m3 wirecolor: [255,255,0]
m13.material = newmat[12]
m13.pivot = [0,-66,0]
rotate m13 -72 [1,0,0]
m14 = copy m13 wirecolor: [0,255,0]
m14.material = newmat[11]
g1 = group #(m6, m14)
g1.pivot = [0,-40,0]
rotate g1 -45 [0,0,1]
move g1 [-5.5,2.5,0]
ungroup g1
m5 = copy m6 wirecolor: [255,255,0]
m5.material = newmat[12]
m7 = copy m4 wirecolor:
[255,255,0]
m7.material = newmat[12]
g2 = group #(m5, m7)
g2.pivot = [0,-40,0]
rotate g2 180 [0,1,0]
ungroup g2
m8 = mesh vertices: #([-4,0,-8],[0,-4,-8],[4,0,-8],[0,4,-8],[-4,0,8],[0,-4,8],[4,0,8],[0,4,8]
,[-8,-4,0],[-8,0,-4],[-8,4,0],[-8,0,4],[8,-4,0],[8,0,-4],[8,4,0],[8,0,4]
,[0,-8,-4],[-4,-8,0],[0,-8,4],[4,-8,0],[0,8,-4],[-4,8,0],[0,8,4],[4,8,0]) \
faces: #([1,3,2],[1,4,3],[5,6,7],[5,7,8],[9,11,10],[9,12,11],[13,14,15],[13,15,16],[17,19,18],[17,20,19],[21,22,23],[21,23,24]
,[1,2,17],[17,18,9],[9,10,1],[1,17,9],[10,22,4],[4,1,10],[10,11,22],[22,21,4],[11,5,23],[23,22,11],[11,12,5],[5,8,23]
,[12,18,6],[6,5,12],[12,9,18],[18,19,6],[8,16,24],[24,23,8],[16,15,24],[8,7,16],[15,3,21],[21,24,15],[15,14,3]
,[3,4,21],[14,20,2],[2,3,14],[14,13,20],[20,17,2],[13,7,19],[19,20,13],[13,16,7],[7,6,19]) \
wirecolor: [0,255,0]
m8.material = newmat[11]
rotate m8 45 [1,0,0]
rotate m8 90 [0,0,1]
move m8 [-37.5,0,0]
m9 = copy m8 wirecolor: [0,255,0]
m9.material
= newmat[11]
m9.pivot = [-37.5,-13,-0.5]
rotate m9 72 [1,0,0]
m10 = copy m8 wirecolor: [0,255,0]
m10.material = newmat[11]
m10.pivot = [-37.5,-13,-0.5]
rotate m10 144 [1,0,0]
m11 = copy m8 wirecolor: [0,255,0]
m11.material = newmat[11]
m11.pivot = [-37.5,-13,-0.5]
rotate m11 18 [1,0,0]
rotate m11 90 [0,0,1]
rotate m11 180 [1,0,0]
rotate m11 -18 [1,0,0]
move m11 [0,7,24]
m12 = copy m9 wirecolor: [255,255,0]
m12.material = newmat[12]
m12.pivot = [-37.5,-23.5,15]
rotate m12 -120 [0,0,1]
attach m3 m4
attach m5 m7
attach m6 m13
attach m3 m5
attach m3 m6
attach m8 m9
attach m8 m11
attach m3 m10
attach m12 m14
attach m3 m12
--
for k = 30 to 43 do(
--
adenosine, cytosine, guanine, timidine, uracil, inosine, pseudouracil,
dihydrouracil, methyl inosine, x-circles maximum, w-methyl-2-guanine,
v-methyl-1-guanine
if (seq[k] == nucl[10]) then (ai =
#(1,1,1,1,1,1,1,1,1,1,1,1,1,1,1); axi = #(1,1,1,1,1,1,1,1); ayi =
#(1,1,1,1,1,1,1,1,1,1,1,1,1,1,1); nuclcol = nuclcolor[10]; nc=10)
if
(seq[k] == nucl[1]) then (ai = #(0,0,0,1,1,1,1,1,1,1,1,1,0,1,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol =
nuclcolor[1]; nc=1)
if (seq[k] == nucl[2]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol = nuclcolor[2]; nc=2)
if
(seq[k] == nucl[3]) then (ai = #(0,1,0,1,1,1,1,1,1,1,1,1,0,1,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,0,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol =
nuclcolor[3]; nc=3)
if (seq[k] == nucl[4]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol = nuclcolor[4]; nc=4)
if
(seq[k] == nucl[5]) then (ai = #(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol =
nuclcolor[5]; nc=5)
if (seq[k] == nucl[6]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,1,1,0,1,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol = nuclcolor[6]; nc=6)
if
(seq[k] == nucl[7]) then (ai = #(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol =
nuclcolor[7]; nc=7)
if (seq[k] == nucl[8]) then (ai =
#(0,0,0,1,1,1,1,1,1,1,0,1,0,0,0); axi = #(0,0,1,1,0,0,0,0); ayi =
#(0,1,0,0,0,0,0,1,0,1,0,1,0,0,0); nuclcol = nuclcolor[8]; nc=8)
if
(seq[k] == nucl[9]) then (ai = #(0,0,0,1,1,1,1,1,1,1,1,1,0,1,0); axi =
#(0,0,1,1,0,0,0,0); ayi = #(0,1,0,0,0,0,0,0,0,1,0,1,0,1,0); nuclcol =
nuclcolor[9]; nc=9)
--
element1 = box length:1 width:1 height:1 position:[0,0,-0.5] wirecolor: nuclcol
Converttomesh element1
for i = 1 to 3 do for j = 1 to 5 do (n = i + 3*(j-1)
if ai[n] > 0 then (aa[n] = box length:10 width:10 height:10 position:[10*(i-2),10*(j-5),10*0.5-10] wirecolor: nuclcol
select #(aa[n]); macros.run "Modifier stack" "convert_to_Mesh"
attach element1 aa[n]
element1.material = newmat[nc]))
element2 = copy m8
element3 = copy m3
attach element1 element3
-- first vector
gr1 = group #(element1,trnk)
gr1.pivot = [-37.451, -2.887, 14.138]
rotate gr1 anglecomp1[k] [0, -27.384, -9.84]
if k == 40 then (rotate gr1 -90 [0, 0, 1] ; rotate gr1 -20 [0, 1, 0] )
ungroup gr1
-- second vector
gr2 = #(element1, element2, trnk)
gr2.pivot = [-43.391, -10.855, 7.631]
rotate gr2 anglecomp2[k] [-11.880, -2.245, -8.094]
ungroup gr2
-- translation vector
attach trnk
element1
attach trnk element2
move trnk [0,-50, 0]
trnk.pivot = [-60,0,0]
-- translation angles
rotate trnk 36 [0, 1, 0]
-- rotate trnk 35 [0, 0, 1]
)
--
delete m3
delete m8
trnk.rotation.controller[2].controller.value = 0
-- animate on
-- at time 100 trnk.rotation.controller[2].controller.value = 360
Анимация (1200*800 пикс, 5 Мб)
Обратите внимание, что вращение, при котором третий нуклеотид иРНК движется азотистым основанием вперёд, т.е. так, чтобы можно было соединиться с комплементарным азотистым основанием сайта рибосомы, соответствует движению тРНК антикодоновой петлёй вперёд. Эта деталь косвенно подтверждает цепочку модельных экспериментов. Другая важнейшая деталь - расположение азотистого основания метилинозина напротив запредельной диэфирной связи триплета иРНК. Дело в том, что для этого нужно совпадение следующих параметров:
1. Координата x
2. Координата y
3. Координата z
4. Угол поворота вокруг оси ox
5. Угол поворота вокруг оси oy
6. Угол поворота вокруг оси oz
Случайностью совпадение 6 параметров быть не может. Вероятность совпадения даже одного параметра ничтожно мала, т.е. составляет менее процента, а процент в шестой степени - это уже триллионная часть. Это означает, что механизм трансляции именно такой.
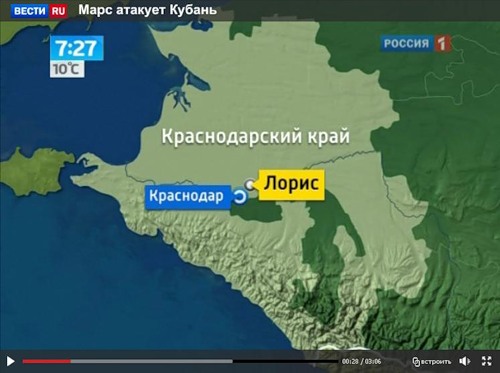
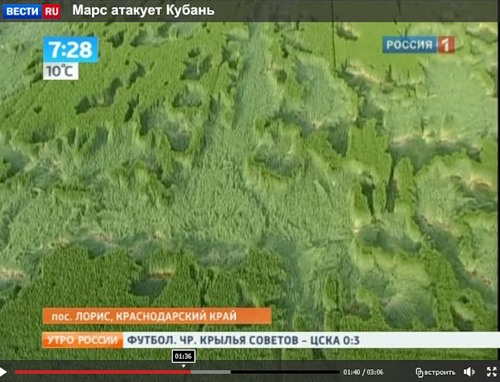
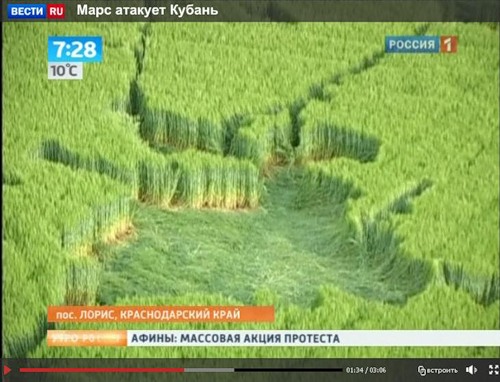

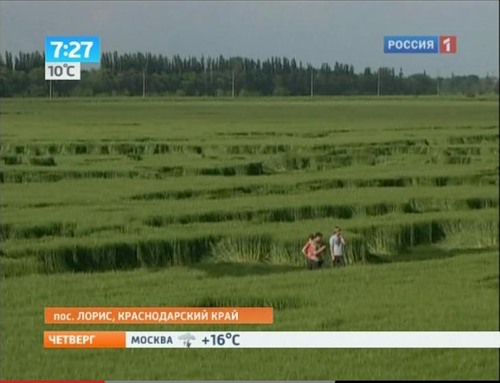
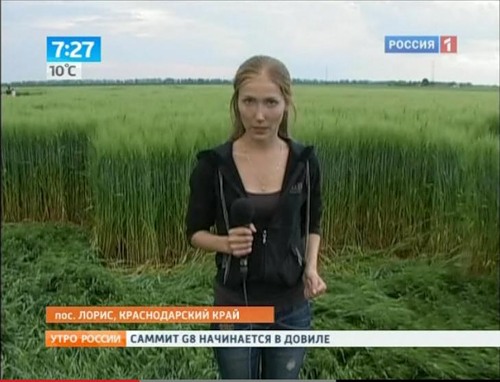
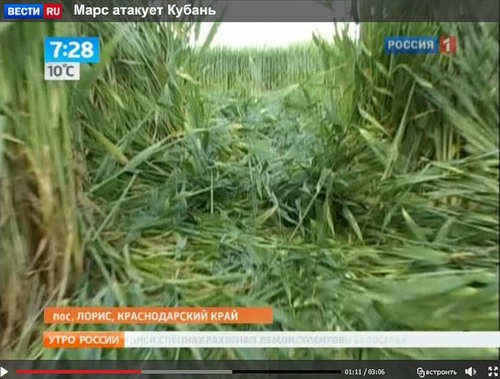

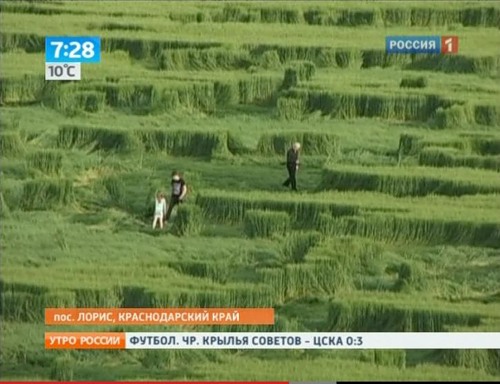
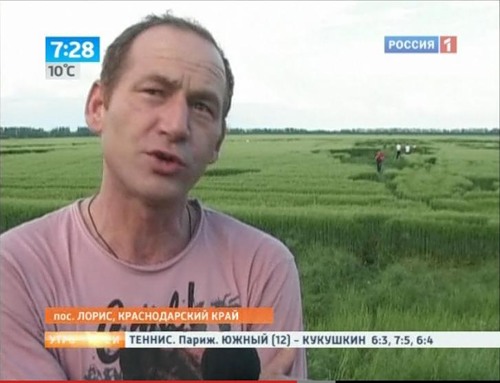
Посмотреть кадры оригинального качества
Аналогичные следы я видел на фотографиях из Англии. Это - типичные следы, которые остаются после сбора урожая драгметаллов инопланетными комбайнами.
Лаборатория Наномир готова к любому взаимовыгодному сотрудничеству. У нас есть сторонники как явные, которые помогают морально и материально, есть очень много пассивных наблюдателей, есть и ярые противники, которые используют любые методы и средства (аморальные и просто преступные), чтобы уничтожить работу лаборатории и дискредитировать ее.
В одиночку внедрить
технологии, выводящие цивилизацию на новый уровень, невозможно.
Благодаря поддержке множества заинтересованных людей проделана огромная
работа. Ознакомиться с её результатами можно изучив материал рассылки "Новости лаборатории Наномир". Люди науки могут изучить научные
труды.
Вклад каждого не останется незамеченным в случае успеха в реализации научных проектов. Результаты совместной деятельности принадлежат участникам проекта пропорционально коэффициентам творческого и финансового участия.
В этом году были куплены рубиновые шарики для эксперимента на сумму ~1000 долл. В результате было сделано научное открытие, проверена защита диэлектрических резонаторов от перенапряжения. В этом же году, вероятно, можно было создать микроволновую энергетику, если бы удалось купить рубиновых шариков на сумму ~5000 долл. или найти сырьё (рубин N11), из которого можно сделать рубиновые шарики для эксперимента в Дубне.
Уже готов эксперимент по созданию "эликсира вечной молодости". Благодаря первому взносу (в размере 500 долларов) Золдракса в ближайшие дни он начнется. Сейчас ведутся переговоры еще с двумя потенциальными инвесторами по поводу финансирования этого проекта.
Созданы первые версии пикотехнологии, с помощью которой Александр Кушелев и Виктория
Соколик сделали более10 научных открытий.
Сотрудничество может быть различным:
- участие в научных дискуссиях на форуме (конструктивное)
- совместное создание коммерческого продукта
- поиск инвесторов
- выступить менеджером по продаже готовых коммерческих продуктов
- конструктивные предложения по продвижению идей лаборатории Наномир
- содействие в проведении экспериментов и т.п.
- написание совместных научных статей и т.п.
- материальный
вклад (денежный или обеспечение оборудованием и материалами)
Пожалуйста, сообщайте о своем вкладе, чтобы мы зачли Вас как партнера лаборатории Наномир.
+7-926-5101703, +7-903-2003424, +7-916-8265031, Skype: Kushelev2009, mail: kushelev2011@yandex.ru
веб-мани: WM-кошелек R426964799301